Simulation¶
Infobiotics Workbench features stochastic simulation capabilities for the models specified in Infobiotics Language.
- The various simulation features can be accessed by activating the IBW Simulation perspective, as below.
- Once the simulation perspective is activated, a dedicated Simulation view will appear and will be docked in the right side of the workbench.
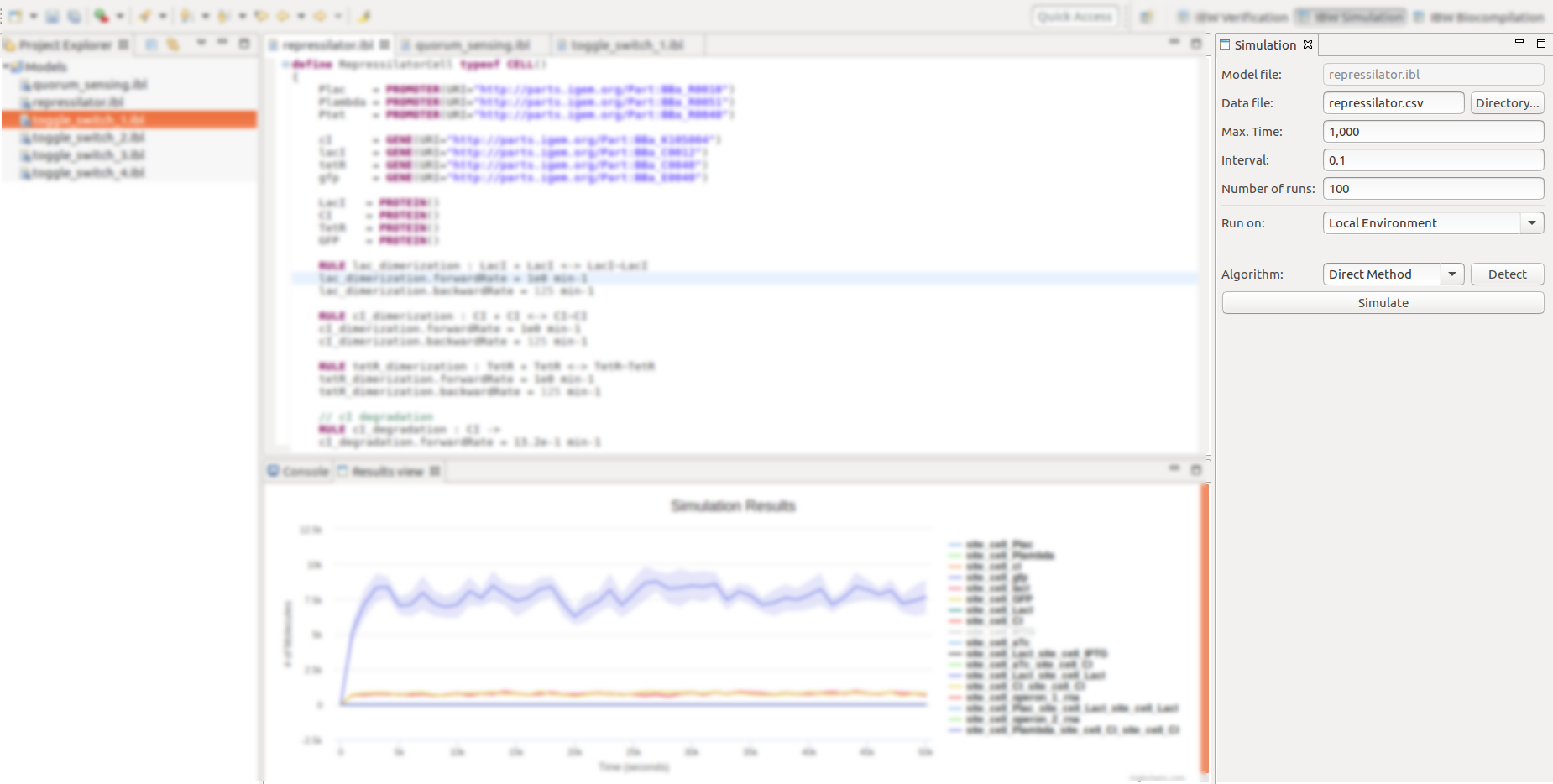
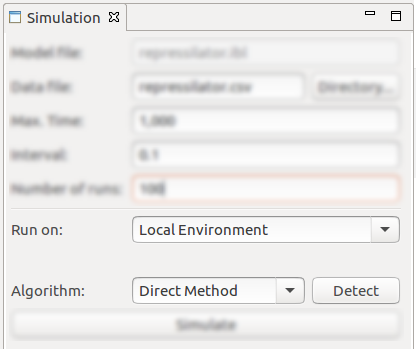
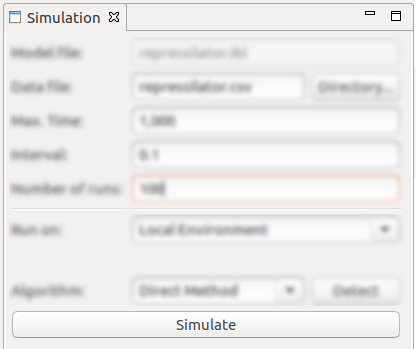
The Simulation view allows the configuration of various parameters and exhibits a button for triggering the actual stochastic simulation:
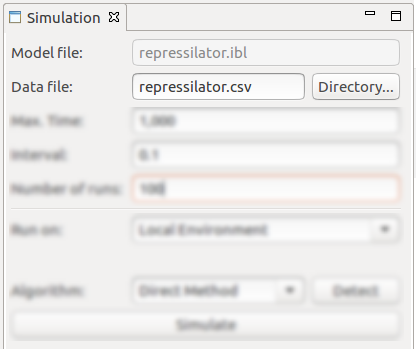
- The location of the csv data file for saving the simulation traces;
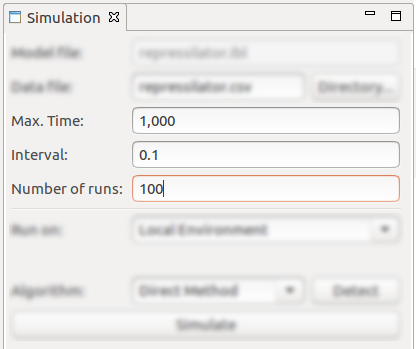
- Max Time, Interval and Number of runs parameters of the stochastic simulation;
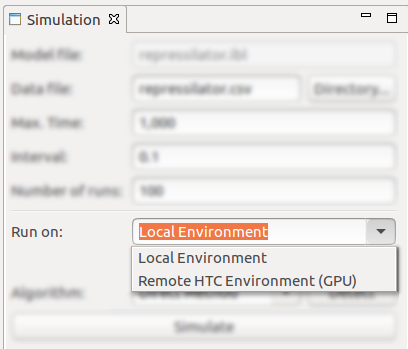
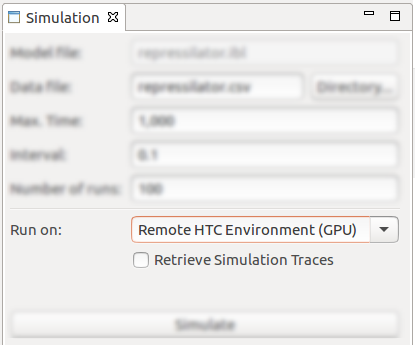
- Environment selection for performing the stochastic simulation. IBW allows the simulation to be performed either locally (on the local machine) or remotely (in a high-performance - GPU computing - environment);
- Performing the simulation on the local environment enables the selection of the algorithm (Gillespie SSA) to be used for stochastic simulation, i.e. Direct Method, First Reaction Method, Next Reaction Method, Optimized Direct Method, Sorting Direct Method, Logarithmic Direct Method, Partial Propensity Direct Method, Composition Rejection, Tau Leaping;
- Performing the simulation on the remove will delegate the simulation to be executed in cloud/on an HPC environment, using a parallel implementation of the Gillespie Logarithmic Direct Method, executing it on GPU. The automated retrieval of the simulation traces from the HPC environment is given as an option;
- After performing the configuration using the above-described parameters, triggering the simulation can be done by clicking on the Simulate button;
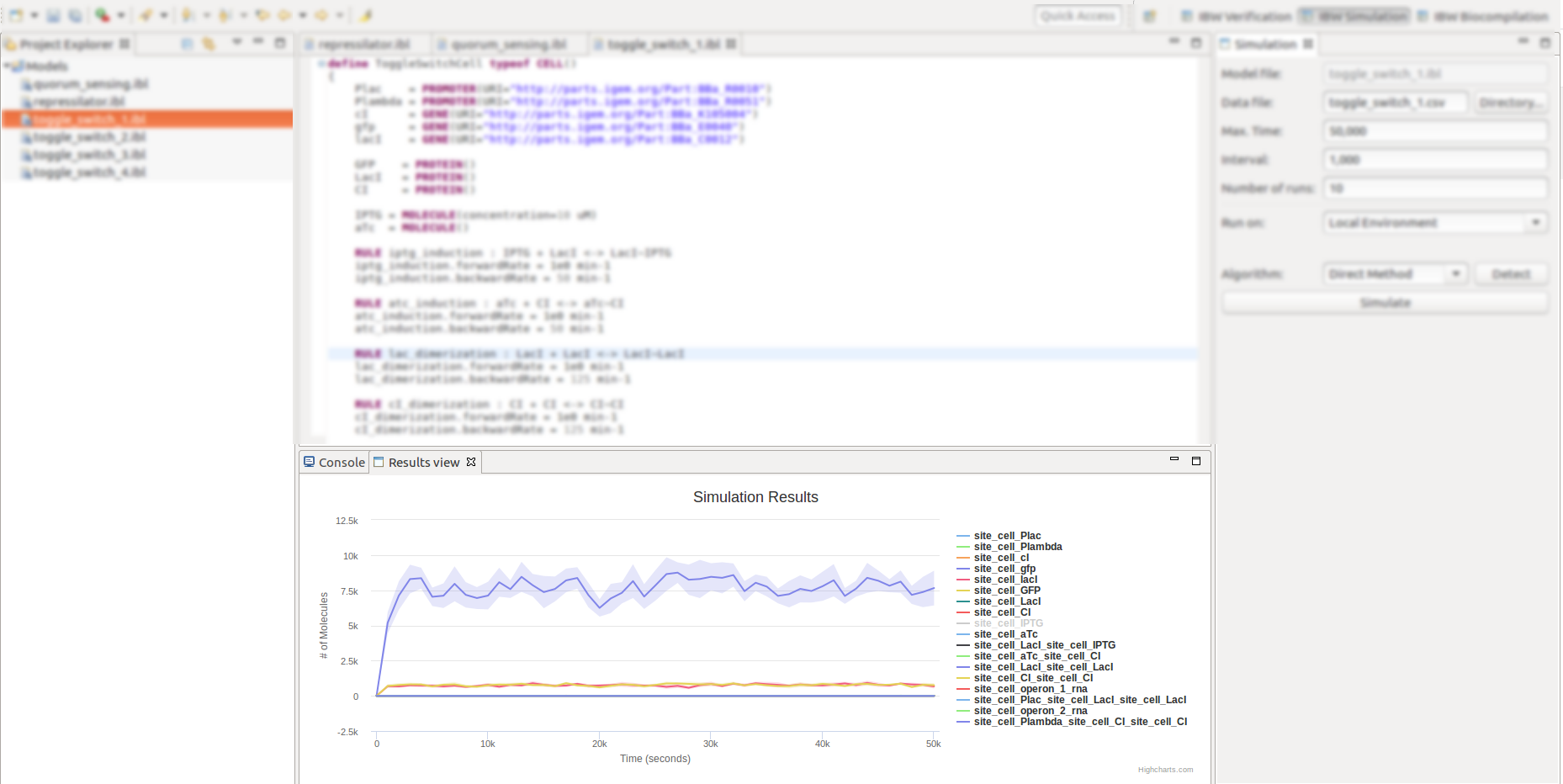
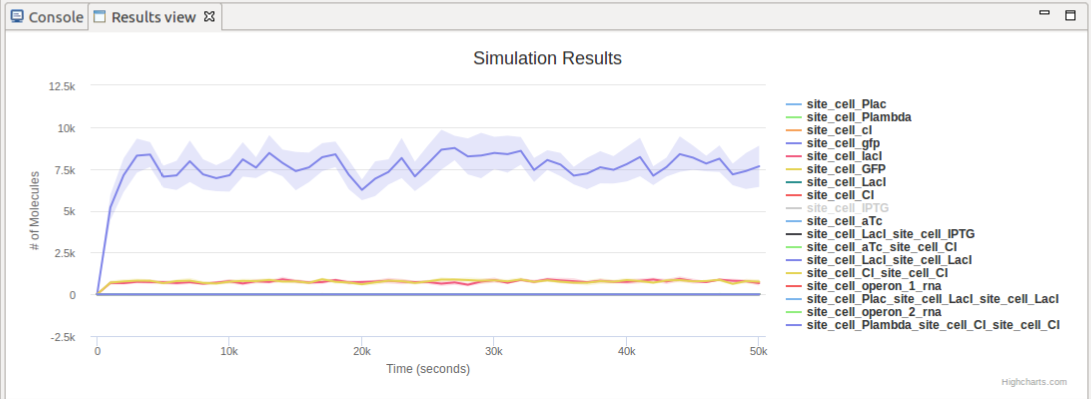
If the simulation has been successfully completed, the Simulation Results view is displayed and docked at the bottom side of the workbench, depicting the simulation traces for the various molecular species, on an interactive chart;