Toggle Switch¶
Intro¶
The Toggle Switch is a seminal synthetic biological circuit that consists of two mutually repressing operons, [Gardner2000] with lacI being regulated by a Plambda promoter and cI (a repressor of Plambda) being in turn regulated by Plac. The operon that codes for CI also codes for green fluorescent protein (GFP). Using this repressor pair allows the toggle switch to be activated by induction via IPTG and deactivated via aTc. The negative feedback cycle ensures that the switch maintain its on-or-off state even when the triggering inducer is washed out. The bistable behaviour of the switch depends on the cooperativity of the repressor proteins - more specifically, on the right choice of regulators and promoters and their correct dimerization.
Below we simulate the toggle switch in four separate scenarios, each one initialized with the average final state of the previous simulation. That is, we first suspended the cell in a region rich in IPTG and simulate its response in CI, LacI, and GFP production over a period of one hour. We then use the average of the total CI, LacI, and GFP concentrations as initial values for CI, LacI, and GFP for simulation of the cell in an IPTG-depleted region. Again, the average final state after one hour is used to initialize the cell state in a region rich in aTc, a.s.o.
IBL Modelling¶
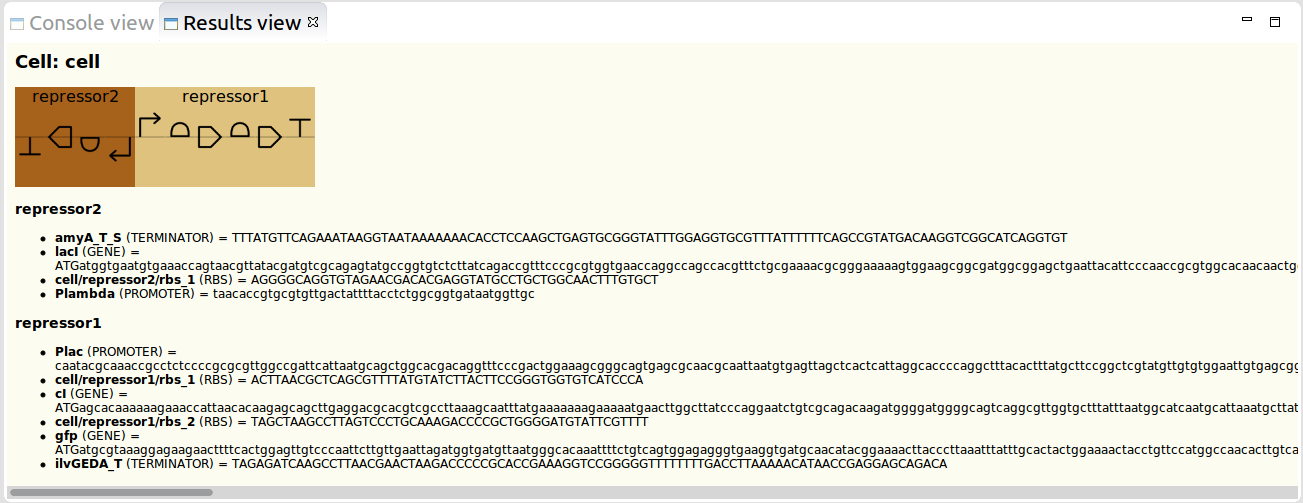
Outlined below are the four Toggle Switch IBL models, corresponding to the four scenarios described above, while the corresponding IBL files can be downloaded here.
Toggle Switch Scenario 1
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 | define ToggleSwitchCell typeof CELL()
{
LacI = PROTEIN()
CI = PROTEIN()
GFP = PROTEIN()
Plac = PROMOTER(URI="http://parts.igem.org/Part:BBa_R0010")
Plambda = PROMOTER(URI="http://parts.igem.org/Part:BBa_R0051")
cI = GENE(URI="http://parts.igem.org/Part:BBa_K105004")
gfp = GENE(URI="http://parts.igem.org/Part:BBa_E0040")
lacI = GENE(URI="http://parts.igem.org/Part:BBa_C0012")
IPTG = MOLECULE(concentration=10 uM)
aTc = MOLECULE()
RULE iptg_induction : IPTG + LacI <-> LacI~IPTG
iptg_induction.forwardRate = 1e0 min-1
iptg_induction.backwardRate = 50 min-1
RULE atc_induction : aTc + CI <-> aTc~CI
atc_induction.forwardRate = 1e0 min-1
atc_induction.backwardRate = 50 min-1
RULE lac_dimerization : LacI + LacI <-> LacI~LacI
lac_dimerization.forwardRate = 1e0 min-1
lac_dimerization.backwardRate = 125 min-1
RULE cI_dimerization : CI + CI <-> CI~CI
cI_dimerization.forwardRate = 1e0 min-1
cI_dimerization.backwardRate = 125 min-1
// cI degradation
RULE cI_degradation : CI ->
cI_degradation.forwardRate = 4.2e-1 min-1
// LacI degradation
RULE lac_degradation : LacI ->
lac_degradation.forwardRate = 13.2e0 min-1
RULE gfp_degradation : GFP ->
gfp_degradation.forwardRate = 6.2e-2 min-1 // 13.216397448e-3 min-1 from Gardner data // 2.5e-2 min-1 // from Andersen et al. Appl Env Microbiol (1998) 64(6):2240-2246
operon_1 = DEVICE(
parts=[Plac,cI,gfp],
input=[LacI~LacI],
output=[CI,GFP]
)
{
rna = RNA()
// repression
RULE repression : Plac + LacI~LacI <-> Plac~LacI~LacI
repression.forwardRate = 1e0 min-1
repression.backwardRate = 10 min-1
// transcription
RULE transcription : Plac -> Plac + rna
transcription.forwardRate = 1.e0 min-1
// translation
RULE translation : rna -> rna + CI + GFP
translation.forwardRate = 100 min-1
// rna degradation
RULE rna_degradation : rna ->
rna_degradation.forwardRate = 1e-1 min-1
}
operon_2 = DEVICE(
parts=[Plambda,lacI],
input=[CI~CI],
output=[LacI]
)
{
rna = RNA()
// repression
RULE repression : Plambda + CI~CI <-> Plambda~CI~CI
repression.forwardRate = 1e0 min-1
repression.backwardRate = 10 min-1
// transcription
RULE transcription : Plambda -> Plambda + rna
transcription.forwardRate = 1.e0 min-1
// translation
RULE translation : rna -> rna + LacI
translation.forwardRate = 100 min-1
// rna degradation
RULE rna_degradation : rna ->
rna_degradation.forwardRate = 1e-1 min-1
ATGC DIRECTION : BACKWARD
}
ATGC ARRANGE Plambda,Plac
VERIFY [ IPTG >= 10uM ] IS FOLLOWED BY [ GFP > 10uM ]
VERIFY [ IPTG >= 10uM ] IS FOLLOWED BY [ GFP > 10uM ] WITH PROBABILITY?
}
define site typeof REGION() {
CELL cell = new ToggleSwitchCell()
}
|
Toggle Switch Scenario 2
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 | define ToggleSwitchCell typeof CELL()
{
LacI = PROTEIN(concentration=0.1 uM)
CI = PROTEIN(concentration=2 uM)
GFP = PROTEIN(concentration=10 uM)
Plac = PROMOTER(URI="http://parts.igem.org/Part:BBa_R0010")
Plambda = PROMOTER(URI="http://parts.igem.org/Part:BBa_R0051")
cI = GENE(URI="http://parts.igem.org/Part:BBa_K105004")
gfp = GENE(URI="http://parts.igem.org/Part:BBa_E0040")
lacI = GENE(URI="http://parts.igem.org/Part:BBa_C0012")
IPTG = MOLECULE()
aTc = MOLECULE()
RULE iptg_induction : IPTG + LacI <-> LacI~IPTG
iptg_induction.forwardRate = 1e0 min-1
iptg_induction.backwardRate = 50 min-1
RULE atc_induction : aTc + CI <-> aTc~CI
atc_induction.forwardRate = 1e0 min-1
atc_induction.backwardRate = 50 min-1
RULE lac_dimerization : LacI + LacI <-> LacI~LacI
lac_dimerization.forwardRate = 1e0 min-1
lac_dimerization.backwardRate = 125 min-1
RULE cI_dimerization : CI + CI <-> CI~CI
cI_dimerization.forwardRate = 1e0 min-1
cI_dimerization.backwardRate = 125 min-1
/// cI degradation
RULE cI_degradation : CI ->
cI_degradation.forwardRate = 4.2e-1 min-1
// LacI degradation
RULE lac_degradation : LacI ->
lac_degradation.forwardRate = 13.2e0 min-1
RULE gfp_degradation : GFP ->
gfp_degradation.forwardRate = 6.2e-2 min-1 // 13.216397448e-3 min-1 from Gardner data // 2.5e-2 min-1 // from Andersen et al. Appl Env Microbiol (1998) 64(6):2240-2246
repressor1 = DEVICE(
parts=[Plac,cI,gfp],
input=[LacI~LacI],
output=[CI,GFP]
)
{
rna = RNA()
// repression
RULE repression : Plac + LacI~LacI <-> Plac~LacI~LacI
repression.forwardRate = 1e0 min-1
repression.backwardRate = 10 min-1
// transcription
RULE transcription : Plac -> Plac + rna
transcription.forwardRate = 1.e0 min-1
// translation
RULE translation : rna -> rna + CI + GFP
translation.forwardRate = 100 min-1
// rna degradation
RULE rna_degradation : rna ->
rna_degradation.forwardRate = 1e-1 min-1
}
repressor2 = DEVICE(
parts=[Plambda,lacI],
input=[CI~CI],
output=[LacI]
)
{
rna = RNA()
// repression
RULE repression : Plambda + CI~CI <-> Plambda~CI~CI
repression.forwardRate = 1e0 min-1
repression.backwardRate = 10 min-1
// transcription
RULE transcription : Plambda -> Plambda + rna
transcription.forwardRate = 1.e0 min-1
// translation
RULE translation : rna -> rna + LacI
translation.forwardRate = 100 min-1
// rna degradation
RULE rna_degradation : rna ->
rna_degradation.forwardRate = 1e-1 min-1
ATGC DIRECTION : BACKWARD
}
ATGC ARRANGE Plambda,Plac
VERIFY [ IPTG >= 0uM ] IS FOLLOWED BY [ GFP/CI > 10] WITH PROBABILITY?
}
define site typeof REGION() {
CELL cell = new ToggleSwitchCell()
}
|
Toggle Switch Scenario 3
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 | define ToggleSwitchCell typeof CELL()
{
LacI = PROTEIN(concentration=0.1 uM)
CI = PROTEIN(concentration=2 uM)
GFP = PROTEIN(concentration=40 uM)
Plac = PROMOTER(URI="http://parts.igem.org/Part:BBa_R0010")
Plambda = PROMOTER(URI="http://parts.igem.org/Part:BBa_R0051")
cI = GENE(URI="http://parts.igem.org/Part:BBa_K105004")
gfp = GENE(URI="http://parts.igem.org/Part:BBa_E0040")
lacI = GENE(URI="http://parts.igem.org/Part:BBa_C0012")
IPTG = MOLECULE()
aTc = MOLECULE(concentration=100 uM)
RULE iptg_induction : IPTG + LacI <-> LacI~IPTG
iptg_induction.forwardRate = 1e0 min-1
iptg_induction.backwardRate = 50 min-1
RULE atc_induction : aTc + CI <-> aTc~CI
atc_induction.forwardRate = 1e0 min-1
atc_induction.backwardRate = 50 min-1
RULE lac_dimerization : LacI + LacI <-> LacI~LacI
lac_dimerization.forwardRate = 1e0 min-1
lac_dimerization.backwardRate = 125 min-1
RULE cI_dimerization : CI + CI <-> CI~CI
cI_dimerization.forwardRate = 1e0 min-1
cI_dimerization.backwardRate = 125 min-1
// cI degradation
RULE cI_degradation : CI ->
cI_degradation.forwardRate = 13.2e0 min-1
// LacI degradation
RULE lac_degradation : LacI ->
lac_degradation.forwardRate = 4.2e-1 min-1
RULE gfp_degradation : GFP ->
gfp_degradation.forwardRate = 6.2e-2 min-1 // 13.216397448e-3 min-1 from Gardner data // 2.5e-2 min-1 // from Andersen et al. Appl Env Microbiol (1998) 64(6):2240-2246
repressor1 = DEVICE(
parts=[Plac,cI,gfp],
input=[LacI~LacI],
output=[CI,GFP]
)
{
rna = RNA()
// repression
RULE repression : Plac + LacI~LacI <-> Plac~LacI~LacI
repression.forwardRate = 1e0 min-1
repression.backwardRate = 10 min-1
// transcription
RULE transcription : Plac -> Plac + rna
transcription.forwardRate = 1.e0 min-1
// translation
RULE translation : rna -> rna + CI + GFP
translation.forwardRate = 100 min-1
// rna degradation
RULE rna_degradation : rna ->
rna_degradation.forwardRate = 1e-1 min-1
}
repressor2 = DEVICE(
parts=[Plambda,lacI],
input=[CI~CI],
output=[LacI]
)
{
rna = RNA()
// repression
RULE repression : Plambda + CI~CI <-> Plambda~CI~CI
repression.forwardRate = 1e0 min-1
repression.backwardRate = 10 min-1
// transcription
RULE transcription : Plambda -> Plambda + rna
transcription.forwardRate = 1.e0 min-1
// translation
RULE translation : rna -> rna + LacI
translation.forwardRate = 100 min-1
// rna degradation
RULE rna_degradation : rna ->
rna_degradation.forwardRate = 1e-1 min-1
ATGC DIRECTION : BACKWARD
}
ATGC ARRANGE Plambda,Plac
VERIFY [GFP] EVENTUALLY DECREASES WITH PROBABILITY ? GIVEN [aTc=100 uM]
}
define site typeof REGION() {
CELL cell = new ToggleSwitchCell()
}
|
Toggle Switch Scenario 4
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 | define ToggleSwitchCell typeof CELL()
{
LacI = PROTEIN(concentration=2 uM)
CI = PROTEIN(concentration=0.1 uM)
GFP = PROTEIN(concentration=23 uM)
Plac = PROMOTER(URI="http://parts.igem.org/Part:BBa_R0010")
Plambda = PROMOTER(URI="http://parts.igem.org/Part:BBa_R0051")
cI = GENE(URI="http://parts.igem.org/Part:BBa_K105004")
gfp = GENE(URI="http://parts.igem.org/Part:BBa_E0040")
lacI = GENE(URI="http://parts.igem.org/Part:BBa_C0012")
IPTG = MOLECULE()
aTc = MOLECULE()
RULE iptg_induction : IPTG + LacI <-> LacI~IPTG
iptg_induction.forwardRate = 1e0 min-1
iptg_induction.backwardRate = 50 min-1
RULE atc_induction : aTc + CI <-> aTc~CI
atc_induction.forwardRate = 1e0 min-1
atc_induction.backwardRate = 50 min-1
RULE lac_dimerization : LacI + LacI <-> LacI~LacI
lac_dimerization.forwardRate = 1e0 min-1
lac_dimerization.backwardRate = 125 min-1
RULE cI_dimerization : CI + CI <-> CI~CI
cI_dimerization.forwardRate = 1e0 min-1
cI_dimerization.backwardRate = 125 min-1
// cI degradation
RULE cI_degradation : CI ->
cI_degradation.forwardRate = 13.2e0 min-1
// LacI degradation
RULE lac_degradation : LacI ->
lac_degradation.forwardRate = 4.2e-1 min-1
RULE gfp_degradation : GFP ->
gfp_degradation.forwardRate = 6.2e-2 min-1 // 13.216397448e-3 min-1 from Gardner data // 2.5e-2 min-1 // from Andersen et al. Appl Env Microbiol (1998) 64(6):2240-2246
repressor1 = DEVICE(
parts=[Plac,cI,gfp],
input=[LacI~LacI],
output=[CI,GFP]
)
{
rna = RNA()
// repression
RULE repression : Plac + LacI~LacI <-> Plac~LacI~LacI
repression.forwardRate = 1e0 min-1
repression.backwardRate = 10 min-1
// transcription
RULE transcription : Plac -> Plac + rna
transcription.forwardRate = 1.e0 min-1
// translation
RULE translation : rna -> rna + CI + GFP
translation.forwardRate = 100 min-1
// rna degradation
RULE rna_degradation : rna ->
rna_degradation.forwardRate = 1e-1 min-1
}
repressor2 = DEVICE(
parts=[Plambda,lacI],
input=[CI~CI],
output=[LacI]
)
{
rna = RNA()
// repression
RULE repression : Plambda + CI~CI <-> Plambda~CI~CI
repression.forwardRate = 1e0 min-1
repression.backwardRate = 10 min-1
// transcription
RULE transcription : Plambda -> Plambda + rna
transcription.forwardRate = 1.e0 min-1
// translation
RULE translation : rna -> rna + LacI
translation.forwardRate = 100 min-1
// rna degradation
RULE rna_degradation : rna ->
rna_degradation.forwardRate = 1e-1 min-1
ATGC DIRECTION : BACKWARD
}
ATGC ARRANGE Plambda,Plac
}
define site typeof REGION() {
CELL cell = new ToggleSwitchCell()
}
|
Simulation¶
In order to analyse the evolution in the concentration of GFP (red), CI (green) and LacI (orange), stochastic simulation has been performed for each of the four scenarios presented above, using the following parameters:
- Max Time: 1000 seconds;
- Interval: 10 seconds;
- Number of runs: 50;
- Gillespie algorithm: Direct Method.
Toggle Switch Scenario 1
The cell is suspended in IPTG, leading to the activation of the switch and production of GFP and CI.
Toggle Switch Scenario 2
CI and GFP production is maintained, despite the absence of IPTG.
Toggle Switch Scenario 3
The cell is suspended in aTc, leading to the deactivation of the switch, production of LacI and decay of GFP.
Toggle Switch Scenario 4
The switch resides in its off state, despite the absence of aTc.
Verification¶
The properties below have been verified in order to validate the system behaviour. The verification has been performed using the embedded Prism and MC2 model checkers.
- The suspension of the cell with IPTG will lead to the production of GFP;
- Once the cell is suspended with IPTG, the GFP concentration will eventually exceed 10uM, with a probability of 79%;
- GFP is produced at least 10 times more than CI, despite the absence of IPTG, with a probability of 49%;
- GFP production will decline if the cell is suspended with aTc.
| # | Verification Statement | Result |
| 1 | VERIFY [IPTG >= 10 uM] IS FOLLOWED BY [GFP > 10 uM] | True |
| 2 | VERIFY [IPTG >= 10 uM] IS FOLLOWED BY [GFP > 10 uM] WITH PROBABILITY ? | 79% |
| 3 | VERIFY [IPTG = 0 uM] IS FOLLOWED BY [GFP/CI > 10] WITH PROBABILITY ? | 49% |
| 4 | VERIFY [GFP] EVENTUALLY DECREASES WITH PROBABILITY ? GIVEN [aTc = 100 uM] | 100% |