Importing and Exporting¶
Infobiotics Workbench features import and export capabilities between Infobiotics Language models and the community-approved standards SBML and SBOL. IBL models can be exported to both SBML and SBOL, while only SBOL imports are allowed, at this moment.
SBOL conversion captures the structural information and hierarchy of the IBL model. This hierarchy (approximately region->cell->molecule/device) is translated into SBOL objects, and if the model is further biocompiled, biocompilation specific information (sequences, cloning sites, and ribosome-binding sites) is also retained. However, SBOL omits numerical information like concentrations, rate constants, compartment sizes, and units. This prevents the SBOL model from being simulated and verified.
SBML conversion captures functional aspects of the IBL model, which includes the previously mentioned numerical information. SBML also retains the structural hierarchy, although biocompilation-specific features, like DNA structures, cannot be expressed.
- Exporting
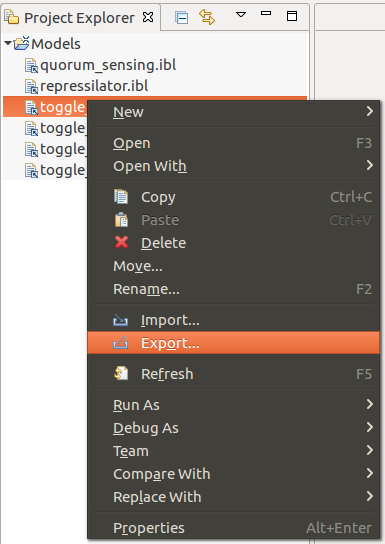
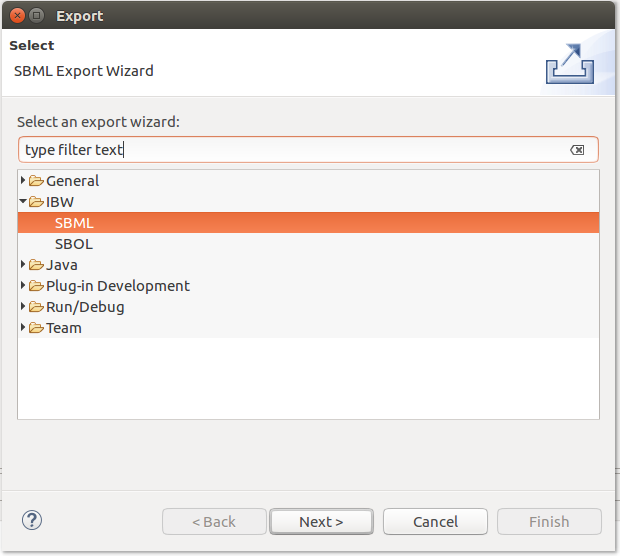
The Infobiotics Language models can be exported by accessing the Export option from the contextual menu of an IBL file, as depicted below:
- Importing
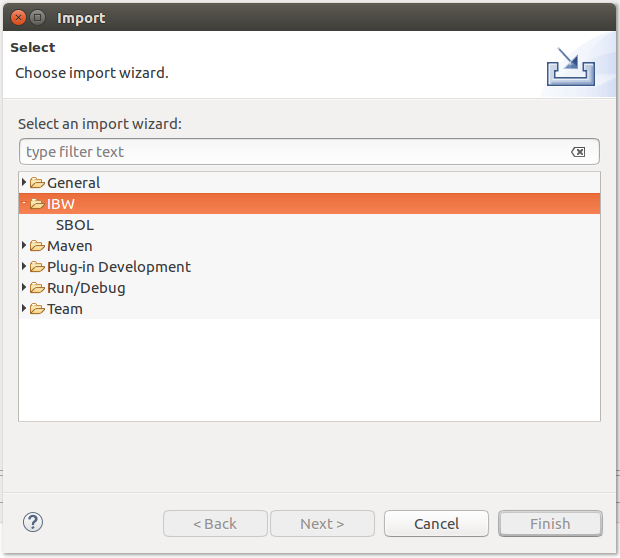
SBOL models can be imported by accessing the Import option from the contextual menu of a selected project directory, as depicted below: