The Repressilator¶
Intro¶
The Repressilator is a seminal early-implemented synthetic genetic circuit, developed by Michael B. Elowitz and Stanislas Leibler [Elowitz2000].
The Repressilator consists of three genes, codifying three repressors. Namely, the operon lactose repressor, lacI, the repressor form the tetracycline transposon, tetR, and a repressor from the λ phage virus, cI. These genes are connected in a synthetic gene regulatory network such that LacI represses the expression of the tetR gene, which in turn represses the cI gene. Finally, the cycle is closed as CI represses the expression of the lacI gene. Thus, the repressilator consists of mutually repressing repressors and, as a result, none of the promoter activation states remains stable and the operons sequentially activate and inactivate each other - resulting in continued global oscillations.
IBL Modelling¶
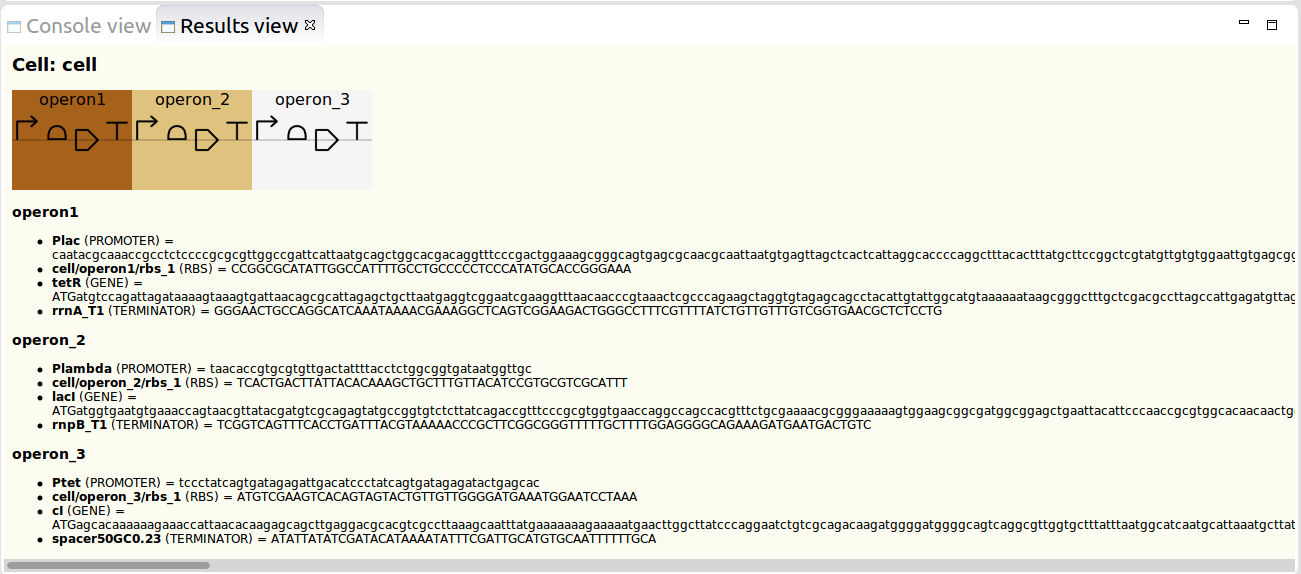
Outlined below is the IBL model for the Repressilator circuit, while the corresponding IBL file can be downloaded here.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 | define RepressilatorCell typeof CELL()
{
LacI = PROTEIN()
CI = PROTEIN()
TetR = PROTEIN()
GFP = PROTEIN()
Plac = PROMOTER(URI="http://parts.igem.org/Part:BBa_R0010")
Plambda = PROMOTER(URI="http://parts.igem.org/Part:BBa_R0051")
Ptet = PROMOTER(URI="http://parts.igem.org/Part:BBa_R0040")
cI = GENE(URI="http://parts.igem.org/Part:BBa_K105004")
lacI = GENE(URI="http://parts.igem.org/Part:BBa_C0012")
tetR = GENE(URI="http://parts.igem.org/Part:BBa_C0040")
gfp = GENE(URI="http://parts.igem.org/Part:BBa_E0040")
RULE lac_dimerization : LacI + LacI <-> LacI~LacI
lac_dimerization.forwardRate = 1e0 min-1
lac_dimerization.backwardRate = 125 min-1
RULE cI_dimerization : CI + CI <-> CI~CI
cI_dimerization.forwardRate = 1e0 min-1
cI_dimerization.backwardRate = 125 min-1
RULE tetR_dimerization : TetR + TetR <-> TetR~TetR
tetR_dimerization.forwardRate = 1e0 min-1
tetR_dimerization.backwardRate = 125 min-1
// cI degradation
RULE cI_degradation : CI ->
cI_degradation.forwardRate = 13.2e-1 min-1
// LacI degradation
RULE lac_degradation : LacI ->
lac_degradation.forwardRate = 13.2e-1 min-1
// TetR degradation
RULE tetR_degradation : TetR ->
tetR_degradation.forwardRate = 13.2e-1 min-1
// GFP degradation
RULE gfp_degradation : GFP ->
gfp_degradation.forwardRate = 13.2e-2 min-1 // 13.216397448e-3 min-1 from Gardner data // 2.5e-2 min-1 // from Andersen et al. Appl Env Microbiol (1998) 64(6):2240-2246
operon1 = DEVICE(
parts=[Plac,tetR],
input=[LacI~LacI],
output=[TetR]
)
{
rna = RNA()
// repression
RULE repression : Plac + LacI~LacI <-> Plac~LacI~LacI
repression.forwardRate = 1e0 min-1
repression.backwardRate = 10 min-1
// transcription
RULE transcription : Plac -> Plac + rna
transcription.forwardRate = 1.e0 min-1
// translation
RULE translation : rna -> rna + TetR
translation.forwardRate = 100 min-1
// rna degradation
RULE rna_degradation : rna ->
rna_degradation.forwardRate = 1e-1 min-1
}
operon_2 = DEVICE(
parts=[Plambda,lacI],
input=[CI~CI],
output=[LacI]
)
{
rna = RNA()
// repression
RULE repression : Plambda + CI~CI <-> Plambda~CI~CI
repression.forwardRate = 1e0 min-1
repression.backwardRate = 10 min-1
// transcription
RULE transcription : Plambda -> Plambda + rna
transcription.forwardRate = 1.e0 min-1
// translation
RULE translation : rna -> rna + LacI
translation.forwardRate = 100 min-1
// rna degradation
RULE rna_degradation : rna ->
rna_degradation.forwardRate = 1e-1 min-1
}
operon_3 = DEVICE(
parts=[Ptet,cI],
input=[TetR~TetR],
output=[CI]
) {
rna = RNA()
// repression
RULE repression : Ptet + TetR~TetR <-> Ptet~TetR~TetR
repression.forwardRate = 1e0 min-1
repression.backwardRate = 10 min-1
// transcription
RULE transcription : Ptet -> Ptet + rna
transcription.forwardRate = 1.e0 min-1
// translation
RULE translation : rna -> rna + CI
translation.forwardRate = 100 min-1
// rna degradation
RULE rna_degradation : rna ->
rna_degradation.forwardRate = 1e-1 min-1
}
VERIFY [LacI <= 1.2 uM] HOLDS INFINITELY OFTEN WITH PROBABILITY ?
VERIFY [LacI > 1.2 uM] HOLDS INFINITELY OFTEN WITH PROBABILITY ?
VERIFY [[CI >= 0.1 uM] AND [CI < 2 uM]] HOLDS IN STEADY-STATE WITH PROBABILITY ?
}
define site typeof REGION() {
CELL cell = new RepressilatorCell()
}
|
Simulation¶
In order to analyse the trends in the concentration of the various molecular species, stochastic simulation has been performed, using the following parameters:
- Max Time: 97,200 seconds (27 hours);
- Interval: 500 seconds;
- Number of runs: 5;
- Gillespie algorithm: Tau Leaping
As depicted in the screenshot below, the result of the stochastic simulation confirms continued oscillations of the regulating proteins LacI (yellow), CI (green) and TetR (red):
Verification¶
Given below is a series of properties that have been used for validating the system behaviour:
- The LacI concentration repeatedly falls below 1.25 uM, with a probability of 56%;
- The LacI concentration repeatedly rises over 1.25 uM, with a probability of 61%;
- The steady state concentration of CI is between 0.1 uM and 2 uM, with a probability of 96%.
| # | Verification Statement | Result |
| 1 | VERIFY [LacI <= 1.25 uM] HOLDS INFINITELY OFTEN WITH PROBABILITY ? | 0.56 |
| 2 | VERIFY [LacI > 1.25 uM] HOLDS INFINITELY OFTEN WITH PROBABILITY ? | 0.61 |
| 3 | VERIFY [[CI >= 0.1 uM] AND [CI < 2 uM]] HOLDS IN STEADY-STATE WITH PROBABILITY ? | 0.96 |